We are a research group at the Guangzhou National Laboratory. We are a research group at the Guangzhou Laboratory. If you are interested in working with us, please see more information on (Vacancies).
RNA centre (Miao lab) is a computational biology laboratory, which focuses on the research of RNA structure, function ( RNA structural informatics ) and single-cell omics sequencing. We develop new computaitonal approaches (algorithms, databases, integrated computational workflows) to understand the RNA function at regulation level and structure level.
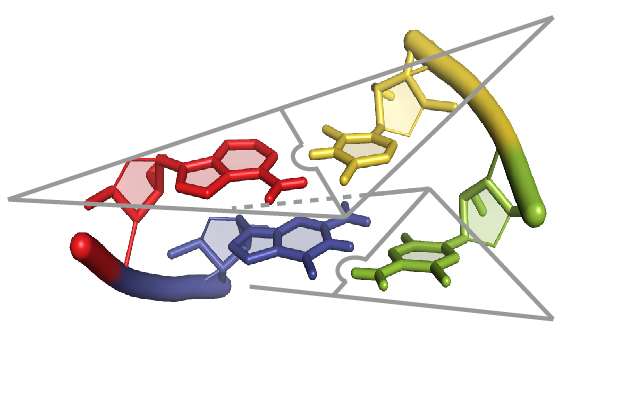
RNA structural informatics:
Our lab seeks an agile and predictive understanding of how RNAs code for information processing and replication in living systems. We are creating new computational and chemical tools to enable the precise modeling and design of these RNAs.
single-cell omics:
We have a longstanding interest in understanding global principles of gene regulation and protein-RNA interactions. We use state-of-the-art genomics approaches, including multi-modal single cell genomics and spatial genomics in combination with machine learning methods to advance our knowledge of cells and tissues.
We are looking for passionate Associate Investigators, Postdocs, Assistant Investigators and Research Assistants to join the team (more info) !
We are grateful for funding from Guangzhou Laboratory (Gzlab), MOST and NSFC .

9. Jun 2025
Yiting Fang will join the lab as a Master student.
9. Jun 2025
Yijun Chen will join the lab as a Master student.
16. May 2025
Bohan Wang is joining the lab as a Research Assistant.
14. Apr 2025
Wenjili Tian is joining the lab as a Research Assistant.
18. Mar 2025
We published a paper in Angewandte Chemie From Theophylline to Adenine or preQ1: Repurposing a DNA Aptamer Revealed by Crystal Structure Analysis. This study elucidated the DNA theophylline aptamer’s ligand-binding mechanism through high-resolution crystal structures of its complexes with theophylline, 3-methylxanthine, and hypoxanthine, and provided a relatively simple and effective method for developing new aptamers.
10. Jan 2025
Our research team organized an unforgettable team-building trip to Nankunshan, more details are here.
30. Dec 2024
We published a paper in Nature Communications Alleviating batch effects in cell type deconvolution with SCCAF-D. We present ‘SCCAF-D’ (Single-Cell Clustering Assessment Framework optimised reference for Deconvolution), a framework that achieves stable accuracies about 0.75 Pearson Correlation Coefficient (PCC) in a ‘cross-reference’ setting via optimally prepared reference data, thereby improving the accuracy of cell proportion inference.
30. Dec 2024
We published a paper in European Journal of Human Genetics RICTOR variants are associated with neurodevelopmental disorders. This study revealed that the mTORC2 pathway was hyperactivated in a patient’s fibroblasts carrying a missense variant, while the expression of RICTOR remained unchanged, indicating a gain-of-function mechanism and confirming the potential role of RICTOR in neuronal cell development.
2. Dec 2024
Recently, Prof.Zhichao Miao and Academician Eric Westhof collaborated on an article entitled RNA-Puzzles Round V:Blind predictions of 23 RNA structures in the Nature Methods. The article published the results of RNA-Puzzles Round V, which conducted a large-scale evaluation of predictions from 18 teams around the world, involving 23 RNA structures, including RNA elements, aptamers, viral elements, ribozymes, and nuclear switches.