Our lab will mainly use computational approaches combined with experiments to understand RNA virus and RNA structure at gene expression regulation and structural levels. Specifically, we use RNA structure prediction and RNA design to udnerstand and engineer RNA molecules. We are hosting the RNA-Puzzles, which is a world-wide community efforts in evaluation of RNA 3D structure prediction. We explore RNA structural biology using a combination of X-ray crystallography, chemical probing, artificial intelligence and computational modeling. Besides, we use single-cell multi-omics to understand the human immune response to RNA viruses, neuron development and immune cell development. We develop computational algorithms to deal with key bottlenecks in single-cell analysis (e.g.,batch effect, cell type clustering, cell type deconvolution). We also develop cloud computing platform and big data atlases for single-cell omics.
Here are some themes that we currently work on (RNA-Puzzles, RNA design, Single-cell omics algorithms, Single-cell omics data analysis ):
It is a world-wide community experiment for blind RNA structure prediction.
It aims to:
The image is a screenshot of our RNA-Puzzles webpage.
We aim to design catalytic RNA molecule as well as RNA inhibitors using the RNA structure folding rules. It is widely known that RNA form base-pairs and secondary structures. However, the “base-pairs” are normally referring to cis-Watson-Crick base-pairs of A-U or G=C, sometimes G-U, while the non-Watson-Crick base-pairs also play a key role in RNA structure formation. We aim to integrate experimental information and RNA structural rules derived from X-ray crystal structures to optimise RNA design including the non-Watson-Crick base-pairs.
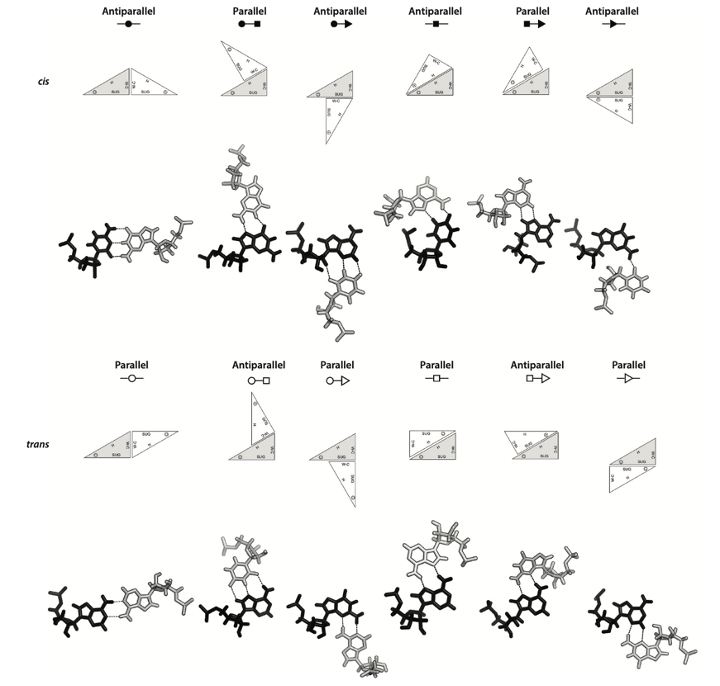
The image is a description about all the base-pair conformations from our Annual Reveiew of Biophysics paper.
Single-cell multi-omics and spatial technologies now provide an opportunity to dissect the state of individual cells at unprecedented resolution. Given the rapid development of different single-cell technologies and new wave of big omics data with their own specificities in terms of complexity and information content, our aims is to develop new computational and theoretical approaches to deal with big single-cell omics data to derive novel biological insights. We aim to understand the spatial and temporal cellular response to RNA viruses.

The image is a cover picture of our SCCAF algorithm.
We use computational analysis to understand the bioligcal significance bebind single-cell omics data. Especially, we are interested in human immune response to RNA viruses, neuron development and immune cell development. We welcome collaborators around but also beyond these topics.
The image is a cover picture of our single-cell research on neuron stem cell in adult primates.