(# co-first author, * corresponding author)
(For a full list of publications and patents see below or go to Google Scholar, ORCID)
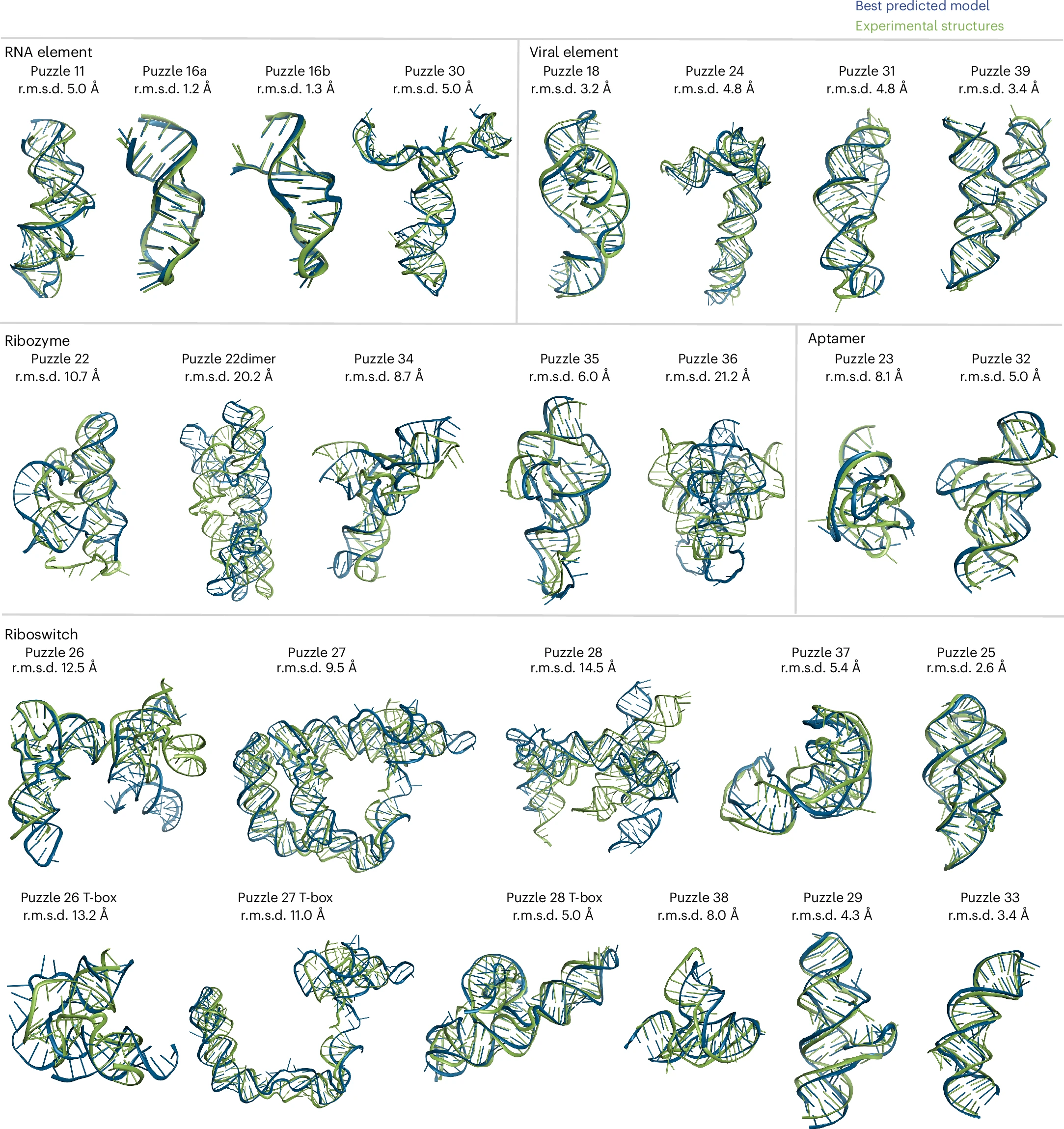
Bu F, Adam Y, Adamiak RW, Antczak M, de Aquino BRH, Badepally NG, Batey RT, Baulin EF, Boinski P, Boniecki MJ, Bujnicki JM, Carpenter KA, Chacon J, Chen SJ, Chiu W, Cordero P, Das NK, Das R, Dawson WK, DiMaio F, Ding F, Dock-Bregeon AC, Dokholyan NV, Dror RO, Dunin-Horkawicz S, Eismann S, Ennifar E, Esmaeeli R, Farsani MA, Ferré-D’Amaré AR, Geniesse C, Ghanim GE, Guzman HV, Hood IV, Huang L, Jain DS, Jaryani F, Jin L, Joshi A, Karelina M, Kieft JS, Kladwang W, Kmiecik S, Koirala D, Kollmann M, Kretsch RC, Kurciński M, Li J, Li S, Magnus M, Masquida B, Moafinejad SN, Mondal A, Mukherjee S, Nguyen THD, Nikolaev G, Nithin C, Nye G, Pandaranadar Jeyeram IPN, Perez A, Pham P, Piccirilli JA, Pilla SP, Pluta R, Poblete S, Ponce-Salvatierra A, Popenda M, Popenda L, Pucci F, Rangan R, Ray A, Ren A, Sarzynska J, Sha CM, Stefaniak F, Su Z, Suddala KC, Szachniuk M, Townshend R, Trachman RJ 3rd, Wang J, Wang W, Watkins A, Wirecki TK, Xiao Y, Xiong P, Xiong Y, Yang J, Yesselman JD, Zhang J, Zhang Y, Zhang Z, Zhou Y, Zok T, Zhang D, Zhang S, Żyła A, Westhof E*, Miao Z*
Nat Methods. (2024). doi:10.1038/s41592-024-02543-9.
We presents a comprehensive single-cell atlas of the human brain, covering nearly all major brain regions in health and disease, and demonstrates the utility of using the atlas to discover putative neural progenitor cells (NPCs) and understand microenvironment-driven differences in microglia. The brain cell atlas will become a valuable resource for studying brain cells and functions, and enhance our understanding of neuronal processes and neurodegenerative diseases.
Xinyue Chen, Yin Huang, Liangfeng Huang, Ziliang Huang, Zhao-Zhe Hao, Lahong Xu, Nana Xu, Zhi Li, Yonggao Mou, Mingli Ye, Renke You, Xuegong Zhang, Sheng Liu*, Zhichao Miao*
Nat. Med. (2024). doi:10.1038/s41591-024-03150-z
We comprehensively mapped the cellular landscape of the primate visual cortex, revealing conservation and diversity of different cell types across species, and identified novel marker genes and cell types using whole-cell electrophysiology, patch clamp and single-cell sequencing (Patch-seq) and fluorescence in situ hybridization.
Wei, J.#, Hao, Z.#, Xu, C., Huang, M., Tang, L., Xu, N., Liu, R., Shen, Y., Teichmann, S. A., Miao, Z.* , & Liu, S.
Nat Commun 13, 6902 (2022). https://doi.org/10.1038/s41467-022-34590-1
Hao, Z.-Z., Wei, J.-R., Xiao, D., Liu, R., Xu, N., Tang, L., Huang, M., Shen, Y., Xing, C., Huang, W., Liu, X., Xiang, M., Liu, Y., Miao, Z.* & Liu, S.*
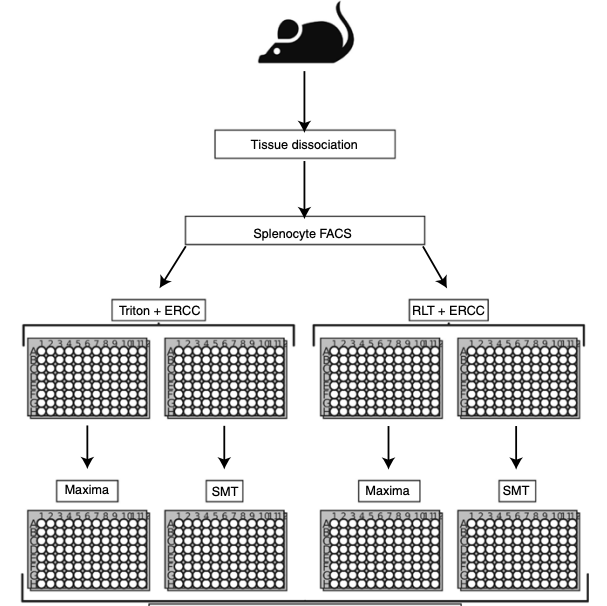
We describe an automated protocol for full-length single-cell RNA sequencing, including both an in-house automated Smart-seq2 protocol and a commercial kit-based workflow. Besidse, we also provide a single-cell sequencing qaulity assessment program SCQUA.
Mamanova, L.*#, Miao, Z.#, Jinat, A., Ellis, P., Shirley, L. & Teichmann, S. A.*
Nat. Protoc. 16, 2886–2915 (2021).

We present the Single-Cell Clustering Assessment Framework (SCCAF), a method for the automated identification of putative cell types from single-cell RNA sequencing (scRNA-seq) data.
Miao, Z., Moreno, P., Huang, N., Papatheodorou, I., Brazma, A.* & Teichmann, S. A.*
Nat. Methods 17, 621–628 (2020).
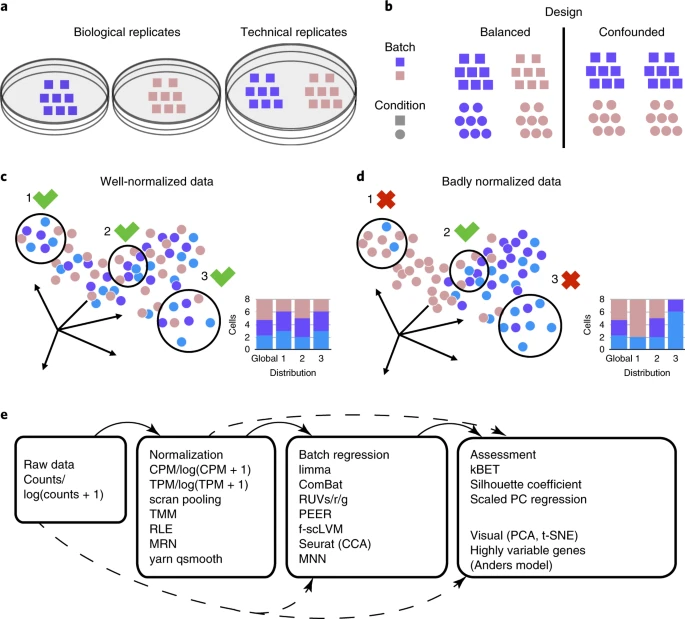
we present a user-friendly, robust and sensitive k-nearest-neighbor batch-effect test (kBET) for quantification of batch effects. We used kBET to assess commonly used batch-regression and normalization approaches, and to quantify the extent to which they remove batch effects while preserving biological variability.
Büttner, M.#, Miao, Z.#, Wolf, F. A., Teichmann, S. A.* & Theis, F. J.*
Nat. Methods 16, 43–49 (2019).
Miao, Z., Adamiak, R. W., Antczak, M., Batey, R. T., Becka, A. J., Biesiada, M., Boniecki, M. J., Bujnicki, J. M., Chen, S.-J., Cheng, C. Y., Chou, F.-C., Ferré-D’Amaré, A. R., Das, R., Dawson, W. K., Ding, F., Dokholyan, N. V., Dunin-Horkawicz, S., Geniesse, C., Kappel, K., Kladwang, W., Krokhotin, A., Łach, G. E., Major, F., Mann, T. H., Magnus, M., Pachulska-Wieczorek, K., Patel, D. J., Piccirilli, J. A., Popenda, M., Purzycka, K. J., Ren, A., Rice, G. M., Santalucia, J., Jr, Sarzynska, J., Szachniuk, M., Tandon, A., Trausch, J. J., Tian, S., Wang, J., Weeks, K. M., Williams, B., 2nd, Xiao, Y., Xu, X., Zhang, D., Zok, T. & Westhof, E.*
highlighted by Faculty Opinions
Ribocentre-aptamer: an integrative, structure-focused database for RNA aptamers
Zhizhong Lu, Hao Sun, Bo Fu, Ying Ao, Jiali Wang, Ke Chen, Yuhang Luo, Linfei Li, Zhaoji Qiu, Jiaxin Zhao, Yuxuan Sun, Zhijie Tan, Qiaozhen Liu, Yangyi Ren, Ziyu Guo, Xiaoxue Chen, Yuanlin He, Yingying Zhao, Yixin Chen, Mengxiao Li, Xuemei Peng, Baowei Huang, Shuang Zhu*, Lin Huang*, Zhichao Miao*
Nucleic acids research, gkaf1016. (2025).
The RNA-Puzzles Assessments of RNA-Only Targets in CASP16
Eric Westhof*, Hao Sun, Fan Bu, Zhichao Miao*
Proteins, 10.1002/prot.70052. (2025).
Enhancing RNA 3D Structure Prediction: A Hybrid Approach Combining Expert Knowledge and Computational Tools in CASP16
Bowen Xiao, Yaohuang Shi, Lin Huang
Proteins, 10.1002/prot.70034. (2025).
The transcription factor Bcl11a is essential for B-1a cell maintenance during aging
Shasha Xu, Liangfeng Huang, Xingjie Liu, Linlin Zhang, Jing Wang, Yifeng Hu, Yi Yang, Xiaojie Shi, Chaohong Liu, Rong Wang, Zhichao Miao*, Yong Yu
Proceedings of the National Academy of Sciences of the United States of America vol. 122,27 (2025).
From Theophylline to Adenine or preQ1: Repurposing a DNA Aptamer Revealed by Crystal Structure Analysis
Xiaowei Lin, Yuanyin Huang, Jinchao Huang, Hao Yuan, Yuhang Luo, Zhizhong Lu, Ying Ao, Jian Huang, Shuo-Bin Chen*, Zhichao Miao*, Lin Huang*
Angewandte Chemie. (2025). doi:10.1002/anie.202504107.
RICTOR variants are associated with neurodevelopmental disorders
Carapito R, Molitor A, Pavinato L, Skeyni A, Lambert M, Pichot A, Jiang J, Spinnhirny P, Zimmermann L, Boucher P, Chung CWT, Elserafy N, Blair EM, Li D, Elisabeth B, Kotzaeridou U, Karch S, Wagner M, Lunsing RJ, Pfundt R, Boycott KM, Bruel AL, Mau-Them FT, Moutton S, Conti V, Mei D, Cetica V, Guerrini R, Brunet T, Rump P, Mussa A, Brusco A, Lemire G, de Vries BBA, Miao Z, Isidor B, Bahram S
Eur J Hum Genet. (2024). doi:10.1038/s41431-024-01774-w.
Alleviating batch effects in cell type deconvolution with SCCAF-D
Feng S, Huang L, Pournara AV, Huang Z, Yang X, Zhang Y, Brazma A, Shi M, Papatheodorou I, Miao Z*
Nat Commun. (2024). doi:10.1038/s41467-024-55213-x.
RNA-Puzzles Round V: blind predictions of 23 RNA structures
Bu F, Adam Y, Adamiak RW, Antczak M, de Aquino BRH, Badepally NG, Batey RT, Baulin EF, Boinski P, Boniecki MJ, Bujnicki JM, Carpenter KA, Chacon J, Chen SJ, Chiu W, Cordero P, Das NK, Das R, Dawson WK, DiMaio F, Ding F, Dock-Bregeon AC, Dokholyan NV, Dror RO, Dunin-Horkawicz S, Eismann S, Ennifar E, Esmaeeli R, Farsani MA, Ferré-D’Amaré AR, Geniesse C, Ghanim GE, Guzman HV, Hood IV, Huang L, Jain DS, Jaryani F, Jin L, Joshi A, Karelina M, Kieft JS, Kladwang W, Kmiecik S, Koirala D, Kollmann M, Kretsch RC, Kurciński M, Li J, Li S, Magnus M, Masquida B, Moafinejad SN, Mondal A, Mukherjee S, Nguyen THD, Nikolaev G, Nithin C, Nye G, Pandaranadar Jeyeram IPN, Perez A, Pham P, Piccirilli JA, Pilla SP, Pluta R, Poblete S, Ponce-Salvatierra A, Popenda M, Popenda L, Pucci F, Rangan R, Ray A, Ren A, Sarzynska J, Sha CM, Stefaniak F, Su Z, Suddala KC, Szachniuk M, Townshend R, Trachman RJ 3rd, Wang J, Wang W, Watkins A, Wirecki TK, Xiao Y, Xiong P, Xiong Y, Yang J, Yesselman JD, Zhang J, Zhang Y, Zhang Z, Zhou Y, Zok T, Zhang D, Zhang S, Żyła A, Westhof E*, Miao Z*
Nat Methods. (2024). doi:10.1038/s41592-024-02543-9.
Virome analysis provides new insights into the pathogenesis mechanism and treatment of SLE disease
Wu Y, Zhang Z, Wang X, Liu X, Qiu Y, Ge X, Miao Z, Meng X, Peng Y
Front Cell Infect Microbiol. (2024).
A pleiotropic recurrent dominant ITPR3 variant causes a complex multisystemic disease
Molitor, A., Lederle, A., Radosavljevic, M., Sapuru, V., Me, Z. T., Yang, J., Shirin, M., Collin-Bund, V., Jerabkova-Roda, K., Miao, Z., Bernard, A., Rolli, V., Grenot, P., Castro, C. N., Rosenzwajg, M., Lewis, E. G., Person, R., Esperón-Moldes, U. S., Kaare, M., Nokelainen, P. T., Batzir, N. A., Hoffer, G. Z., Paul, N., Stemmelen, T., Naegely, L., Hanauer, A., Bibi-Triki, S., Grün, S., Jung, S., Busnelli, I., Tripolszki, K., Al-Ali, R., Ordonez, N., Bauer, P., Song, E., Zajo, K., Partida-Sanchez, S., Robledo-Avila, F., Kumanovics, A., Louzoun, Y., Hirschler, A., Pichot, A., Toker, O., Mejía, C. A. M., Parvaneh, N., Knapp, E., Hersh, J. H., Kenney, H., Delmonte, O. M., Notarangelo, L. D., Goetz, J. G., Kahwash, S. B., Carapito, C., Bajwa, R. P. S., Thomas, C., Ehl, S., Isidor, B., Carapito, R., Abraham, R. S., Hite, R. K., Marcus, N., Bertoli-Avella, A. & Bahram, S.
Science advances10, (2024).
Loss of histone deubiquitinase Bap1 triggers anti-tumor immunity
Chang, H., Li, M., Zhang, L., Li, M., Ong, S. H., Zhang, Z., Zheng, J., Xu, X., Zhang, Y., Wang, J., Liu, X., Li, K., Luo, Y., Wang, H., Miao, Z., Chen, X., Zha, J. & Yu, Y.
Cell. Oncol. (2024). doi:10.1007/s13402-024-00978-y
A cellular reference atlas across human brain regions
Liangfeng Huang, Zhichao Miao*
Nat. Med. 1–2 (2024).
A brain cell atlas integrating single-cell transcriptomes across human brain regions
Xinyue Chen, Yin Huang, Liangfeng Huang, Ziliang Huang, Zhao-Zhe Hao, Lahong Xu, Nana Xu, Zhi Li, Yonggao Mou, Mingli Ye, Renke You, Xuegong Zhang, Sheng Liu*, Zhichao Miao*
Nat. Med. (2024). doi:10.1038/s41591-024-03150-z
CTCF mutation at R567 causes developmental disorders via 3D genome rearrangement and abnormal neurodevelopment
Jie Zhang, Gongcheng Hu, Yuli Lu, Huawei Ren, Yin Huang, Yulin Wen, Binrui Ji, Diyang Wang, Haidong Wang, Huisheng Liu, Ning Ma, Lingling Zhang, Guangjin Pan, Yibo Qu, Hua Wang, Wei Zhang, Zhichao Miao, Hongjie Yao*
Nat. Commun. 15, (2024).
Clinical genomics expands the link between erroneous cell division, primary microcephaly and intellectual disability
Saima, Amjad Khan*, Sajid Ali, Jiuhong Jiang, Zhichao Miao, Atif Kamil, Shahid Niaz Khan, Stefan T Arold
Neurogenetics (2024). doi:10.1007/s10048-024-00759-7
CATD: a reproducible pipeline for selecting cell-type deconvolution methods across tissues
Anna Vathrakokoili Pournara* , Zhichao Miao* , Ozgur Yilimaz Beker, Nadja Nolte, Alvis Brazma, Irene Papatheodorou*
Bioinformatics Advances 4, vbae048 (2024).
Exome sequencing identifies homozygous variants in MBOAT7 associated with neurodevelopmental disorder
Nazmina, G., Khan, A.* , Jiang, J. , Miao, Z. , Khan, SN., Khan, MI., Shah, AH., Shah, AH., Khisroon, M.*, Haack, TB.
Clin. Genet. (2023). doi:10.1111/cge.14469
Regulatory circular RNAs in viral diseases: applications in diagnosis and therapy.
Wang, W., Sun, L., Huang, M., Quan, Y., Jiang, T., Miao, Z.* , Zhang, Q.*
RNA Biol. (2023). doi:10.1080/15476286.2023.2272118
Assessment of three-dimensional RNA structure prediction in CASP15.
Das, R.* , Kretsch, RC., Simpkin, AJ., Mulvaney, T., Pham, P., Rangan, R., Bu, F., Keegan, RM., Topf, M., Rigden, DJ.,Miao, Z.* , Westhof, E.*
Proteins (2023). doi:10.1002/prot.26602
Nr4a1 marks a distinctive ILC2 activation subset in the mouse inflammatory lung.
Xu, S., Zhang, Y., Liu, X., Liu, H., Zou, X., Zhang, L., Wang, J., Zhang, Z., Xu, X., Li, M., Li, K., Shi, S., Zhang, Y., Miao, Z., Zha, J., Yu, Y.
BMC Biol. 21, (2023).
Benchmarking strategies for cross-species integration of single-cell RNA sequencing data.
Song, Y., Miao, Z., Brazma, A., Papatheodorou, I.
Nat. Commun. 14, (2023).
Ribocentre-switch: a database of riboswitches.
Bu, F.#, Lin, X.#, Liao, W.#, Lu, Z., He, Y., Luo, Y., Peng, X., Li, M., Huang, Y., Chen, X., Xiao, B., Jiang, J., Deng, J., Huang, J., Lin, T., Miao, Z.* , Huang, L.*
Nucleic Acids Res. (2023). doi:10.1093/nar/gkad891
Single Cell Analysis of the Fate of Injected Oncogenic RasV12 Cells in Adult Wild Type Drosophila.
Chen, D., Lan, X., Huang, X., Huang, J., Zhou, X., Miao, Z., Ma, Y., Goto, A., Ji, S., & Hoffmann, J. A.
J. Innate Immun. 15, (2023).
Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Jia, X., Pan, Z., Yuan, Y., Luo, B., Luo, Y., Mukherjee, S., Jia, G., Liu, L., Ling, X., Yang, X., Miao, Z., Wei, X., Bujnicki, J.M., Zhao, K., Su, Z.
Cell Res. (2023). doi:10.1038/s41422-023-00786-3
Sterile 20-like kinase 3 promotes tick-borne encephalitis virus assembly by interacting with NS2A and prM and enhancing the NS2A-NS4A association.
Tang, J., Xu, C., Fu, M., Liu, C., Zhang, X., Zhang, W., Pei, R., Wang, Y., Zhou, Y., Chen, J., Miao, Z., Pan, G., Yang, Q., Chen, X.
J. Med. Virol. (2023). doi:10.1002/jmv.28610
RNA 3D Structure Comparison Using RNA-Puzzles Toolkit
Magnus, M., & Miao, Z.*
RNA Structure Prediction 263–285 (2023).
Defects and asymmetries in the visual pathway of non-human primates with natural strabismus and amblyopia
Liu, F., Wang, Z., Huang, W., Xu, Y., Sang, X., Liu, R., Li, Z., Bi, Y., Tang, L., Peng, J., Wei, J., Miao, Z., Yan, J., & Liu, S.
Zool Res 44, 153–168 (2023).
Identification of visual cortex cell types and species differences using single-cell RNA sequencing
Wei, J.#, Hao, Z.#, Xu, C., Huang, M., Tang, L., Xu, N., Liu, R., Shen, Y., Teichmann, S. A., Miao, Z.* , & Liu, S.
Nat Commun 13, 6902 (2022). https://doi.org/10.1038/s41467-022-34590-1
Ribocentre: a database of ribozymes.
Deng, J.#, Shi, Y.#, Peng, X.#, He, Y., Chen, X., Li, M., Lin, X., Liao, W., Huang, Y., Jiang, T., Lilley, D. M. J., Miao, Z.* , & Huang, L.*
Nucleic Acids Res. gkac840 (2022).
Single-cell atlas of human liver development reveals pathways directing hepatic cell fates.
Wesley, B. T.#, Ross, A. D. B.#, Muraro, D., Miao, Z., Saxton, S., Tomaz, R. A., Morell, C. M., Ridley, K., Zacharis, E. D., Petrus-Reurer, S., Kraiczy, J., Mahbubani, K. T., Brown, S., Garcia-Bernardo, J., Alsinet, C., Gaffney, D., Horsfall, D., Tysoe, O. C., Botting, R. A., Stephenson, E., Popescu, D.-M., MacParland, S., Bader, G., McGilvray, I. D., Ortmann, D., Sampaziotis, F., Saeb-Parsy, K., Haniffa, M., Stevens, K. R., Zilbauer, M., Teichmann, S. A., & Vallier, L.*
Nat. Cell Biol. (2022). doi:10.1038/s41556-022-00989-7
Linking transcriptomes with morphological and functional phenotypes in mammalian retinal ganglion cells.
Huang, W., Xu, Q., Su, J., Tang, L., Hao, Z.-Z., Xu, C., Liu, R., Shen, Y., Sang, X., Xu, N., Tie, X., Miao, Z., Liu, X., Xu, Y., Liu, F., Liu, Y., & Liu, S.*
Cell Rep. 40, 111322 (2022).
Computational Pipeline for Reference-Free Comparative Analysis of RNA 3D Structures Applied to SARS-CoV-2 UTR Models. International Journal of Molecular Sciences.
Gumna J., Antczak M., Adamiak RW., Bujnicki JM., Chen S-J., Ding F., Ghosh P., Li J., Mukherjee S., Nithin C., Pachulska-Wieczorek K., Ponce-Salvatierra A., Popenda M., Sarzynska J., Wirecki T., Zhang D., Zhang S., Zok T., Westhof E., Miao Z.* , Szachniuk M., & Rybarczyk A.*
Int. J. Mol. Sci. 23, 9630 (2022).
Single-cell transcriptomics of adult macaque hippocampus reveals neural precursor cell populations.
Hao, Z.-Z., Wei, J.-R., Xiao, D., Liu, R., Xu, N., Tang, L., Huang, M., Shen, Y., Xing, C., Huang, W., Liu, X., Xiang, M., Liu, Y., Miao, Z.* & Liu, S.*
Nat. Neurosci. 1–13 (2022).
Identification of TPBG-Expressing Amacrine Cells in DAT-tdTomato Mouse.
Huang, W., Xu, Q., Liu, F., Su, J., Xiao, D., Tang, L., Hao, Z.-Z., Liu, R., Xiang, K., Bi, Y., Miao, Z., Liu, X., Liu, Y. & Liu, S.*
Invest. Ophthalmol. Vis. Sci. 63, 13–13 (2022).
Single-cell genomics identifies distinct B1 cell developmental pathways and reveals aging-related changes in the B-cell receptor repertoire.
Luo, Y., Wang, J., Li, K., Li, M., Xu, S., Liu, X., Zhang, Z., Xu, X., Zhang, Y., Pan, J., Liu, P., Gao, S., Miao, Z.* & Yu, Y.*
Cell Biosci. 12, 57 (2022).
Expression Atlas update: gene and protein expression in multiple species.
Moreno, P., Fexova, S., George, N., Manning, J. R., Miao, Z., Mohammed, S., Muñoz-Pomer, A., Fullgrabe, A., Bi, Y., Bush, N., Iqbal, H., Kumbham, U., Solovyev, A., Zhao, L., Prakash, A., García-Seisdedos, D., Kundu, D. J., Wang, S., Walzer, M., Clarke, L., Osumi-Sutherland, D., Tello-Ruiz, M. K., Kumari, S., Ware, D., Eliasova, J., Arends, M. J., Nawijn, M. C., Meyer, K., Burdett, T., Marioni, J., Teichmann, S., Vizcaíno, J. A., Brazma, A.* & Papatheodorou, I.*
Nucleic Acids Res. 50, D129–D140 (2022).
Single-Cell Analysis Reveals the Immune Characteristics of Myeloid Cells and Memory T Cells in Recovered COVID-19 Patients With Different Severities.
Li, X.#, Garg, M.#, Jia, T., Liao, Q., Yuan, L., Li, M., Wu, Z., Wu, W., Bi, Y., George, N., Papatheodorou, I., Brazma, A., Luo, H., Fang, S., Miao, Z.* ,Shu, Y.*
Frontiers in Immunology 12, 1664-3224 (2022).
Evaluation of the stereochemical quality of predicted RNA 3D models in the RNA-Puzzles submissions.
Carrascoza, F., Antczak, M., Miao, Z., Westhof, E. & Szachniuk, M.*
RNA (2021). doi:10.1261/rna.078685.121
Meta-analysis of COVID-19 single-cell studies confirms eight key immune responses.
Garg, M.#, Li, X.#, Moreno, P., Papatheodorou, I., Shu, Y., Brazma, A. & Miao, Z.*
Sci. Rep. 11, 20833 (2021).
Mapping Rora expression in resting and activated CD4+ T cells.
Haim-Vilmovsky, L., Henriksson, J., Walker, J. A., Miao, Z., Natan, E., Kar, G., Clare, S., Barlow, J. L., Charidemou, E., Mamanova, L., Chen, X., Proserpio, V., Pramanik, J., Woodhouse, S., Protasio, A. V., Efremova, M., Griffin, J. L., Berriman, M., Dougan, G., Fisher, J., Marioni, J. C., McKenzie, A. N. J. & Teichmann, S. A.*
PLoS One 16, e0251233 (2021).
High-throughput full-length single-cell RNA-seq automation.
Mamanova, L.*#, Miao, Z.#, Jinat, A., Ellis, P., Shirley, L. & Teichmann, S. A.*
Nat. Protoc. 16, 2886–2915 (2021).
User-friendly, scalable tools and workflows for single-cell RNA-seq analysis.
Moreno, P., Huang, N., Manning, J. R., Mohammed, S., Solovyev, A., Polanski, K., Bacon, W., Chazarra, R., Talavera-López, C., Doyle, M. A., Marnier, G., Grüning, B., Rasche, H., George, N., Fexova, S. K., Alibi, M., Miao, Z., Perez-Riverol, Y., Haeussler, M., Brazma, A., Teichmann, S., Meyer, K. B. & Papatheodorou, I.*
Nat. Methods 18, 327–328 (2021).
Longevity, clonal relationship, and transcriptional program of celiac disease-specific plasma cells.
Lindeman, I., Zhou, C., Eggesbø, L. M., Miao, Z., Polak, J., Lundin, K. E. A., Jahnsen, J., Qiao, S.-W., Iversen, R. & Sollid, L. M.*
J. Exp. Med. 218, (2021).
Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics.
Muus, C., Luecken, M. D., Eraslan, G., Sikkema, L., Waghray, A., Heimberg, G., Kobayashi, Y., Vaishnav, E. D., Subramanian, A., Smillie, C., Jagadeesh, K. A., Duong, E. T., Fiskin, E., Triglia, E. T., Ansari, M., Cai, P., Lin, B., Buchanan, J., Chen, S., Shu, J., Haber, A. L., Chung, H., Montoro, D. T., Adams, T., Aliee, H., Allon, S. J., Andrusivova, Z., Angelidis, I., Ashenberg, O., Bassler, K., Bécavin, C., Benhar, I., Bergenstråhle, J., Bergenstråhle, L., Bolt, L., Braun, E., Bui, L. T., Callori, S., Chaffin, M., Chichelnitskiy, E., Chiou, J., Conlon, T. M., Cuoco, M. S., Cuomo, A. S. E., Deprez, M., Duclos, G., Fine, D., Fischer, D. S., Ghazanfar, S., Gillich, A., Giotti, B., Gould, J., Guo, M., Gutierrez, A. J., Habermann, A. C., Harvey, T., He, P., Hou, X., Hu, L., Hu, Y., Jaiswal, A., Ji, L., Jiang, P., Kapellos, T. S., Kuo, C. S., Larsson, L., Leney-Greene, M. A., Lim, K., Litviňuková, M., Ludwig, L. S., Lukassen, S., Luo, W., Maatz, H., Madissoon, E., Mamanova, L., Manakongtreecheep, K., Leroy, S., Mayr, C. H., Mbano, I. M., McAdams, A. M., Nabhan, A. N., Nyquist, S. K., Penland, L., Poirion, O. B., Poli, S., Qi, C., Queen, R., Reichart, D., Rosas, I., Schupp, J. C., Shea, C. V., Shi, X., Sinha, R., Sit, R. V., Slowikowski, K., Slyper, M., Smith, N. P., Sountoulidis, A., Strunz, M., Sullivan, T. B., Sun, D., Talavera-López, C., Tan, P., Tantivit, J., Travaglini, K. J., Tucker, N. R., Vernon, K. A., Wadsworth, M. H., Waldman, J., Wang, X., Xu, K., Yan, W., Zhao, W., Ziegler, C. G. K., NHLBI LungMap Consortium & Human Cell Atlas Lung Biological Network. Miao, Z. as a consortium author in Human Cell Atlas Lung Biological Network.
Nat. Med. 27, 546–559 (2021).
NCKAP1L defects lead to a novel syndrome combining immunodeficiency, lymphoproliferation, and hyperinflammation.
Castro, C. N., Rosenzwajg, M., Carapito, R., Shahrooei, M., Konantz, M., Khan, A., Miao, Z., Groß, M., Tranchant, T., Radosavljevic, M., Paul, N., Stemmelen, T., Pitoiset, F., Hirschler, A., Nespola, B., Molitor, A., Rolli, V., Pichot, A., Faletti, L. E., Rinaldi, B., Friant, S., Mednikov, M., Karauzum, H., Aman, M. J., Carapito, C., Lengerke, C., Ziaee, V., Eyaid, W., Ehl, S., Alroqi, F., Parvaneh, N. & Bahram, S.
J. Exp. Med. 217, (2020).
SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes.
Sungnak, W., Huang, N., Bécavin, C., Berg, M., Queen, R., Litvinukova, M., Talavera-López, C., Maatz, H., Reichart, D., Sampaziotis, F., Worlock, K. B., Yoshida, M., Barnes, J. L. & Human Cell Atlas Lung Biological Network. Miao, Z. as a consortium author in Human Cell Atlas Lung Biological Network.
Nat. Med. 1–7 (2020).
Two Cases of Recessive Intellectual Disability Caused by NDST1 and METTL23 Variants.
Khan, A., Miao, Z., Umair, M., Ullah, A., Alshabeeb, M. A., Bilal, M., Ahmad, F., Rappold, G. A., Ansar, M. & Carapito, R.*
Genes 11, 1021 Preprint at https://doi.org/10.3390/genes11091021 (2020)
Secondary structure of the SARS-CoV-2 5’-UTR.
Miao, Z., Tidu, A., Eriani, G. & Martin, F.*
RNA Biol. 18, 447–456 (2021).
Advances in RNA 3D Structure Modeling Using Experimental Data.
Li, B., Cao, Y., Westhof, E. & Miao, Z.*
Front. Genet. 11, (2020).
RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers.
Miao, Z., Adamiak, R. W., Antczak, M., Boniecki, M. J., Bujnicki, J., Chen, S.-J., Cheng, C. Y., Cheng, Y., Chou, F.-C., Das, R., Dokholyan, N. V., Ding, F., Geniesse, C., Jiang, Y., Joshi, A., Krokhotin, A., Magnus, M., Mailhot, O., Major, F., Mann, T. H., Piątkowski, P., Pluta, R., Popenda, M., Sarzynska, J., Sun, L., Szachniuk, M., Tian, S., Wang, J., Wang, J., Watkins, A. M., Wiedemann, J., Xiao, Y., Xu, X., Yesselman, J. D., Zhang, D., Zhang, Y., Zhang, Z., Zhao, C., Zhao, P., Zhou, Y., Zok, T., Żyła, A., Ren, A., Batey, R. T., Golden, B. L., Huang, L., Lilley, D. M., Liu, Y., Patel, D. J. & Westhof, E.*
RNA 26, 982–995 (2020).
Putative cell type discovery from single-cell gene expression data.
Miao, Z., Moreno, P., Huang, N., Papatheodorou, I., Brazma, A.* & Teichmann, S. A.*
Nat. Methods 17, 621–628 (2020).
RNA-Puzzles toolkit: a computational resource of RNA 3D structure benchmark datasets, structure manipulation, and evaluation tools.
Magnus, M., Antczak, M., Zok, T., Wiedemann, J., Lukasiak, P., Cao, Y., Bujnicki, J. M., Westhof, E., Szachniuk, M. & Miao, Z.*
Nucleic Acids Res. 48, 576–588 (2020).
Decoding human fetal liver haematopoiesis.
Popescu, D.-M., Botting, R. A., Stephenson, E., Green, K., Webb, S., Jardine, L., Calderbank, E. F., Polanski, K., Goh, I., Efremova, M., Acres, M., Maunder, D., Vegh, P., Gitton, Y., Park, J.-E., Vento-Tormo, R., Miao, Z., Dixon, D., Rowell, R., McDonald, D., Fletcher, J., Poyner, E., Reynolds, G., Mather, M., Moldovan, C., Mamanova, L., Greig, F., Young, M. D., Meyer, K. B., Lisgo, S., Bacardit, J., Fuller, A., Millar, B., Innes, B., Lindsay, S., Stubbington, M. J. T., Kowalczyk, M. S., Li, B., Ashenberg, O., Tabaka, M., Dionne, D., Tickle, T. L., Slyper, M., Rozenblatt-Rosen, O., Filby, A., Carey, P., Villani, A.-C., Roy, A., Regev, A., Chédotal, A., Roberts, I., Göttgens, B., Behjati, S., Laurenti, E., Teichmann, S. A.* & Haniffa, M.*
Nature 574, 365–371 (2019).
BBKNN: Fast Batch Alignment of Single Cell Transcriptomes.
Polański, K.#, Park, J.-E.#, Young, M. D.#, Miao, Z., Meyer, K. B. & Teichmann, S. A.*
Bioinformatics (2019). doi:10.1093/bioinformatics/btz625
AbRSA: A robust tool for antibody numbering.
Li, L.#, Chen, S.#, Miao, Z.#, Liu, Y., Liu, X., Xiao, Z.-X.* & Cao, Y.*
Protein Sci. 28, 1524–1531 (2019).
Exome sequencing identifies a novel missense variant in CTSC causing nonsyndromic aggressive periodontitis.
Molitor, A., Prud’homme, T., Miao, Z., Conrad, S., Bloch-Zupan, A., Pichot, A., Hanauer, A., Isidor, B., Bahram, S. & Carapito, R.*
Journal of Human Genetics 64, 689–694 Preprint at https://doi.org/10.1038/s10038-019-0615-3 (2019)
Comparative analysis of sequencing technologies for single-cell transcriptomics.
Natarajan, K. N.#, Miao, Z.#, Jiang, M., Huang, X., Zhou, H., Xie, J., Wang, C., Qin, S., Zhao, Z., Wu, L., Yang, N., Li, B., Hou, Y., Liu, S. & Teichmann, S. A.
Genome Biol. 20, 70 (2019).
A test metric for assessing single-cell RNA-seq batch correction.
Büttner, M.#, Miao, Z.#, Wolf, F. A., Teichmann, S. A.* & Theis, F. J.*
Nat. Methods 16, 43–49 (2019).
Genome-wide analyses reveal the IRE1a-XBP1 pathway promotes T helper cell differentiation by resolving secretory stress and accelerating proliferation.
Pramanik, J., Chen, X., Kar, G., Henriksson, J., Gomes, T., Park, J.-E., Natarajan, K., Meyer, K. B., Miao, Z., McKenzie, A. N. J., Mahata, B.* & Teichmann, S. A.*
Genome Med. 10, 76 (2018).
Evaluation of Protein-Ligand Docking by Cyscore.
Cao, Y., Dai, W. & Miao, Z.
Methods in Molecular Biology 233–243 (2018).
Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features.
Carapito, R., Konantz, M., Paillard, C., Miao, Z., Pichot, A., Leduc, M. S., Yang, Y., Bergstrom, K. L., Mahoney, D. H., Shardy, D. L., Alsaleh, G., Naegely, L., Kolmer, A., Paul, N., Hanauer, A., Rolli, V., Müller, J. S., Alghisi, E., Sauteur, L., Macquin, C., Morlon, A., Sancho, C. S., Amati-Bonneau, P., Procaccio, V., Mosca-Boidron, A.-L., Marle, N., Osmani, N., Lefebvre, O., Goetz, J. G., Unal, S., Akarsu, N. A., Radosavljevic, M., Chenard, M.-P., Rialland, F., Grain, A., Béné, M.-C., Eveillard, M., Vincent, M., Guy, J., Faivre, L., Thauvin-Robinet, C., Thevenon, J., Myers, K., Fleming, M. D., Shimamura, A., Bottollier-Lemallaz, E., Westhof, E., Lengerke, C., Isidor, B. & Bahram, S.*
J. Clin. Invest. 127, 4090–4103 (2017).
RNA Structure: Advances and Assessment of 3D Structure Prediction.
Miao, Z.* & Westhof, E.*
Annu. Rev. Biophys. 46, 483–503 (2017).
RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme.
Miao, Z., Adamiak, R. W., Antczak, M., Batey, R. T., Becka, A. J., Biesiada, M., Boniecki, M. J., Bujnicki, J. M., Chen, S.-J., Cheng, C. Y., Chou, F.-C., Ferré-D’Amaré, A. R., Das, R., Dawson, W. K., Ding, F., Dokholyan, N. V., Dunin-Horkawicz, S., Geniesse, C., Kappel, K., Kladwang, W., Krokhotin, A., Łach, G. E., Major, F., Mann, T. H., Magnus, M., Pachulska-Wieczorek, K., Patel, D. J., Piccirilli, J. A., Popenda, M., Purzycka, K. J., Ren, A., Rice, G. M., Santalucia, J., Jr, Sarzynska, J., Szachniuk, M., Tandon, A., Trausch, J. J., Tian, S., Wang, J., Weeks, K. M., Williams, B., 2nd, Xiao, Y., Xu, X., Zhang, D., Zok, T. & Westhof, E.*
RNA 23, 655–672 (2017).
Quantifying side-chain conformational variations in protein structure.
Miao, Z.* & Cao, Y.*
Sci. Rep. 6, 37024 (2016).
RBscore&NBench: a high-level web server for nucleic acid binding residues prediction with a large-scale benchmarking database.
Miao, Z.* & Westhof, E.*
Nucleic Acids Res. 44, W562–7 (2016).
DRSP: a Structural Database for Single Residue Substitutions in PDB.
Liu, J.#, Miao, Z.#, Li, L., Xiao, Z. & Cao, Y.*
pibb.2016.0056. doi:10.16476/j.pibb.2016.0056
Prediction of nucleic acid binding probability in proteins: a neighboring residue network based score.
Miao, Z. & Westhof, E.*
Nucleic Acids Res. 43, 5340–5351 (2015).
RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures.
Miao, Z., Adamiak, R. W., Blanchet, M.-F., Boniecki, M., Bujnicki, J. M., Chen, S.-J., Cheng, C., Chojnowski, G., Chou, F.-C., Cordero, P., Cruz, J. A., Ferré-D’Amaré, A. R., Das, R., Ding, F., Dokholyan, N. V., Dunin-Horkawicz, S., Kladwang, W., Krokhotin, A., Lach, G., Magnus, M., Major, F., Mann, T. H., Masquida, B., Matelska, D., Meyer, M., Peselis, A., Popenda, M., Purzycka, K. J., Serganov, A., Stasiewicz, J., Szachniuk, M., Tandon, A., Tian, S., Wang, J., Xiao, Y., Xu, X., Zhang, J., Zhao, P., Zok, T. & Westhof, E.*
RNA 21, 1066–1084 (2015).
A Large-Scale Assessment of Nucleic Acids Binding Site Prediction Programs.
Miao, Z. & Westhof, E.*
PLoS Comput. Biol. 11, e1004639 (2015).
Modeling of protein side-chain conformations with RASP.
Miao, Z., Cao, Y. & Jiang, T.*
Methods Mol. Biol. 1137, 43–53 (2014).
RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core.
Anokhina, M., Bessonov, S., Miao, Z., Westhof, E., Hartmuth, K. & Lührmann, R.*
EMBO J. 32, 2804–2818 (2013).
RASP: rapid modeling of protein side chain conformations.
Miao, Z., Cao, Y. & Jiang, T.*
Bioinformatics 27, 3117–3122 (2011).
Improved side-chain modeling by coupling clash-detection guided iterative search with rotamer relaxation.
Cao, Y., Song, L., Miao, Z., Hu, Y., Tian, L. & Jiang, T.*
Bioinformatics 27, 785–790 (2011).